.![]() Advances in facial recognition technology may have useful applications in healthcare. Getty Images
Advances in facial recognition technology may have useful applications in healthcare. Getty Images
Mon 19 June 2023:
Foetal alcohol syndrome is a lifelong condition caused by exposing an unborn baby to alcohol. It’s a pattern of mental, physical and behavioural symptoms seen in some people whose mothers consumed alcohol during pregnancy. Not all prenatal alcohol exposure results in the syndrome; it is the most severe form of a range of effects called foetal alcohol spectrum disorders.
South Africa has the highest reported rates of foetal alcohol spectrum disorders in the world: 111.1 per 1,000 population. The disorders may affect seven million people in the country. The number could be higher because of under-diagnosis.
Foetal alcohol syndrome can’t be reversed. But confirmed diagnosis can have benefits. It can lead to early intervention and therapy (physical, occupational, and speech, among others), and a better understanding from parents and teachers. Diagnosis can also ensure that adults are eligible for social services support.
Clinicians use a range of methods to diagnose foetal alcohol syndrome, including assessing abnormal growth and brain function. A key part of the process is looking at the individual’s facial features. Typical features are small eye openings, a thin upper lip, and a smooth area between the nose and upper lip.
But visual examination of the facial features can be subjective and often depends on the clinician’s experience and expertise. Another challenge arises in low-resource settings when there aren’t many doctors specially trained to do this.
A more objective and standard way to detect foetal alcohol syndrome early would therefore be useful.
One method that’s being used to aid diagnosis is three-dimensional (3D) surfaces produced by devices that scan the face. The technology is costly and complex. Two-dimensional (2D) images are easier to get – it can be done with a digital camera or smartphone – but are not accurate enough for diagnosis.
Our study sought to explore whether it was possible to use normal 2D face images to approximate 3D surfaces of the face. We showed that it was. Our method involved using 3D models that can change their shape based on a variety of real human faces, combined with 3D facial analysis technology.
We argue in our paper that our findings show the technology can improve early detection, intervention and treatment for people affected by foetal alcohol syndrome, particularly in low-resource settings.
We hope to contribute to the global effort to prevent and manage the lifelong consequences of the syndrome and disorders.
How it would work
We constructed a flexible 3D model that can alter its shape based on a variety of real human faces. The changes are guided by statistical patterns learned from a dataset of high-quality 3D scans from 98 individuals. This international open-source dataset was carefully curated to represent different demographic groups.
We didn’t have access to image data of individuals affected by foetal alcohol syndrome. We therefore used 2D and 3D images of individuals without this condition to develop and validate our approach. We nevertheless reasoned that our method should work equally well for any scenario where the model and the test subjects are closely matched.
We then set out to develop and validate a machine learning algorithm for predicting 3D faces of unseen subjects, from their 2D face images only, using our 3D model.
This was a pioneering step in our research, where we aimed to create a “smart” tool that could bring flat images to life in three dimensions. The results of the study were encouraging.
Our 3D-from-2D prediction algorithm performed well in three ways:
- capturing facial variations
- representing unique features
- summarising information of faces from 2D images.
Since we had actual 3D face scans to use for comparison, we were able to calculate the average difference between these scans and the face shapes predicted by our model. This allowed us to measure the error in our fitting, which we found to be in line with other studies.
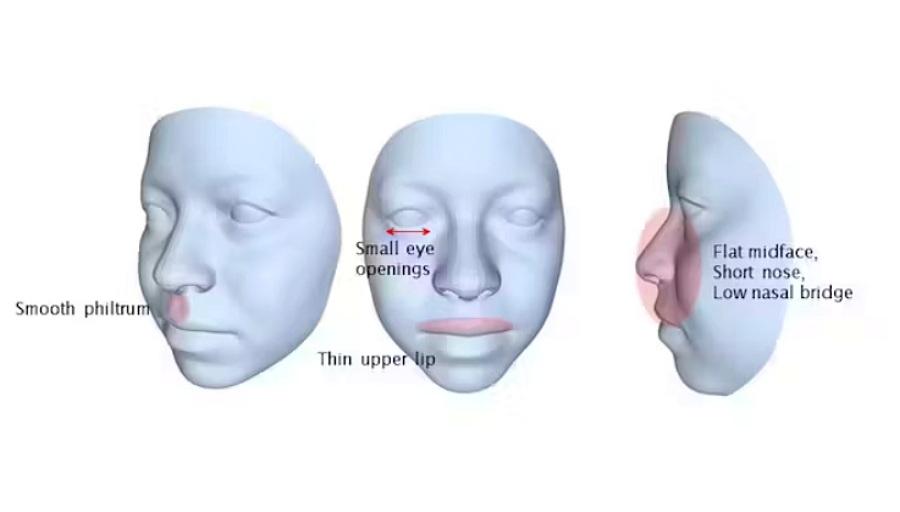
We particularly focused on specific regions of the face: the eyes, midface, upper lip, and philtrum (the groove between the nose and the top lip). These regions provide crucial information for clinicians when examining the facial markers of foetal alcohol syndrome.
We could accurately predict these facial regions, and concluded from this that our method could form the foundation of an image-based diagnostic tool for foetal alcohol syndrome.
Our study also showed that the quality of our predictions was independent of skin tone. This is a crucial finding. Certain 3D scanning technologies have been known to struggle with accurately capturing darker skin tones. This issue is being addressed. Nevertheless, our findings gave us confidence that there was additional potential for use of our approach in diverse populations.
Challenges
We did identify some limitations. Access to 3D data of individuals with foetal alcohol syndrome remains a challenge. Future research could focus on reducing reconstruction errors to acceptable clinical standards by collecting and analysing larger datasets, including data from underrepresented populations.
This Article Originally Published in The Conversation Click Here
![]()
Tinashe Ernest Muzvidzwa Mutsvangwa
Associate Professor of Biomedical Engineering, University of Cape Town
A biomedical engineer and researcher with a PhD in Biomedical Engineering, specializing in medical image analysis and health innovation. As an educator and mentor, actively participating in capacity-building initiatives regionally and globally. Holding roles in various scientific committees and professional societies, and committed to fostering international collaborations and strengthening relationships with scientific organizations. Working on promoting inclusivity, intellectual development, and interdisciplinary collaboration, ultimately impacting society through advancements in healthcare, capacity building, and scientific knowledge exchange.
______________________________________________________________
![]()
Bernhard Egger
Professor for Cognitive Computer Vision, Friedrich-Alexander-Universität Erlangen-Nürnberg
I study how humans and machines can perceive faces and shapes in general. In particular, I choose to focus on statistical shape models and the 3D Morphable Models as generative representation since they naturally disentangle the underlying variables. In combination with computer graphics we simulate the image formation process and approach the inverse problem in an analysis-by-synthesis manner. I’m junior professor at the chair of visual computing at Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU). Before joing FAU I was a postdoc in Josh Tenenbaum’s Computational Cognitive Science Lab at the Departement of Brain and Cognitive Sciences at MIT and the Center for Brains, Minds and Machines (CBMM) and Polina Golland’s group at MIT Computer Science & Artificial Intelligence Lab. I did my PhD on facial image annotation and interpretation in unconstrained images in the Graphics and Vision Research Group at the University of Basel. Before my doctorate I obtained my M.Sc. and B.Sc. in Computer Science at the University of Basel and an upper secondary school teaching Diploma at the University of Applied Sciences Northwestern Switzerland.
______________________________________________________________
![]()
Felix Atuhaire
Lecturer, Mbarara University of Science and Technology
I am currently working as a lecturer in the Department of Biomedical Sciences and Engineering at Mbarara University of Science and Technology. My academic background is in both computer and biomedical engineering.
______________________________________________________________
FOLLOW INDEPENDENT PRESS:
TWITTER (CLICK HERE)
https://twitter.com/IpIndependent
FACEBOOK (CLICK HERE)
https://web.facebook.com/ipindependent
Think your friends would be interested? Share this story!





